
Most of environmental bacteria are motile. They are not only motile but also can sense changes in the concentration of chemicals and respond to them by altering their pattern of motility. They can swim toward “what they like” (attractants), while they swim away from “what they do not like” (repellents). These behavioral responses are called chemotaxis. Environmental bacteria include important plant and animal pathogens, and potential agents of geochemical cycles, biocontrol and bioremediation, and chemotaxis has been thought to play an important role in microbe-host and microbe-substrate interactions because contact with hosts and substrates are essential to start these ecological behaviors. Bacterial chemotaxis has been thought to provide a primitive model for animal sensory system, and most studies have been focused on the genetics and biochemistry of the chemotaxis system in Escherichia coli and Salmonella enterica serovar Typhimurium. But relatively little has been studied on chemotaxis in environmental bacteria. We are interested of chemotaxis in environmental bacteria, especially Pseudomonas species, and have intensively investigated molecular biology of chemotaxis system in Pseudomonas aeruginosa, Pseudomonas putida, Pseudomonas fluorescens and Ralstonia solanaserum to understand the role of chemotaxis in the earliest step of bacterial ecological behaviors and to control them
P. aeruginosa PAO1 as a model microbe for research to understand ecological aspects of chemotaxis*
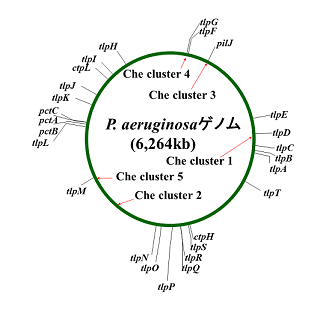
P. aeruginosa inhabits a wide range of environmental niches from soil and water to the human host. Since this bacterium shows chemotactic responses towards various chemical stimuli including 20 naturally-occurred amino acids, sugars, organic acids, inorganic phosphate, oxygen, chloroform, trichloroethylene, and thiocyanic esters. Methods for molecular genetics and biochemistry have been established, and genomic information is fully available, we have studied P. aeruginosa PAO1 as a model microbe for research to understand ecological aspects of chemotaxis. Genomic information reveals that P. aeruginosa possesses 26 putative genes encoding chemotaxis chemosensory proteins and more than 20 putative genes encoding proteins for chemotaxis signal transduction (Fig. 1). We identified genes required for chemotaxis signal transduction and chemotaxis chemosensory proteins for amino acids, inorganic phosphate, oxygen, trichloroethylene, chloroform, ethylene, toluene, and malate. *: J. Biosci. Bioeng. 106:357-362 (2008).
Chemotactic responses to environmental pollutant, trichlorpethylene (TCE) by P. aeruginosa
It is Parales, et al who first reported bacterial chemotaxis to TCE (Appl. Environ. Microbiol. 66:4098-4104 [2000]). They found that P. putida F1 cells grown in the presence of toluene were attracted by TCE and toluene and TodST, two-component regulatory pair for toluene oxidation, were required for chmotaxis to TCE. But, chemotaxis chemosensory protein for TCE has not been identified. We found that P. aeruginosa PAO1 shows negative chemotaxis (repelled response) to TCE. Genetic analysis revealed that chemosensory proteins PctABC, which are also chemosensory proteins for positive chemotaxis to amino acids, are responsible for detection of TCE. In the course of negative chemotaxis to TCE by P. aeruginosa, we obtained unexpected and interesting results. When pctABC deletion mutant of P. aeruginosa PAO1 was tested for chemotaxis to TCE, we found that the mutant unexpectedly showed positive chemotaxis to lower concentrations of TCE. This result clearly demonstrates that P. aeruginosa PAO1 possesses two kinds of chemosensory proteins for TCE, one is for negative chemotaxis and the other is for positive chemotaxis. We examined chemosensory protein mutant collection of P. aeruginosa PAO1 for chemotactic responses to TCE, and found that CttP (PA0180) is a chemosensory protein for positive chemotaxis. In the wild type strain, signal from PctABC is much stronger than that from CttP and the wild type cells exhibit negative chemotaxis to TCE. But, once the PctABC genes are inactivated, the mutant cells show positive chemotaxis to TCE. We are now investigating molecular biology of TCE chemotaxis in P. putida F1. P. putida F1 is capable of degrading TCE in cooxidative manner. If we can enhance its ability to detect and swim to TCE, we can construct “super pollutant degrader with radar function” (Fig. 2).
Chemotaxis to plant-related compounds and its role in the initiation of microbe-plant interactions
We are thinking of chemotaxis as the prelude of ecological behaviors. Especially, we are interested of the role of chemotaxis in the initiation of microbe-plant interactions (for example, infection and symbiosis). We expect that results from the investigation will contribute to development novel technology to prevent bacterial infection to plants and to enhance bacterial symbiosis which has positive effects on plant growth. Plants excrete several compounds into environments. Environmental bacteria should detect such compounds and swim towards plants by using chemotaxis. Major components in plant excretes include amino acids, dicarboxylic acids such as malate and succinate, aromatic compounds, and sugars. First, we investigated chemotaxis of P. aeruginosa to these compounds to identify chemosensory proteins for these compounds. We have identified chemosensory proteins for amino acids, malate, and aromatic compounds. Interestingly, P. aeruginosa was found to respond to ethylene, one of plant hormones. Genetic analysis found that TlpQ serves as a chemosensory protein for ethylene. We are now investigating chemotaxis to plant-related compounds in P. fluorescens (which enhances growth of plants after colonization on plant roots) and R. solanaserum (important plant pathogen).
